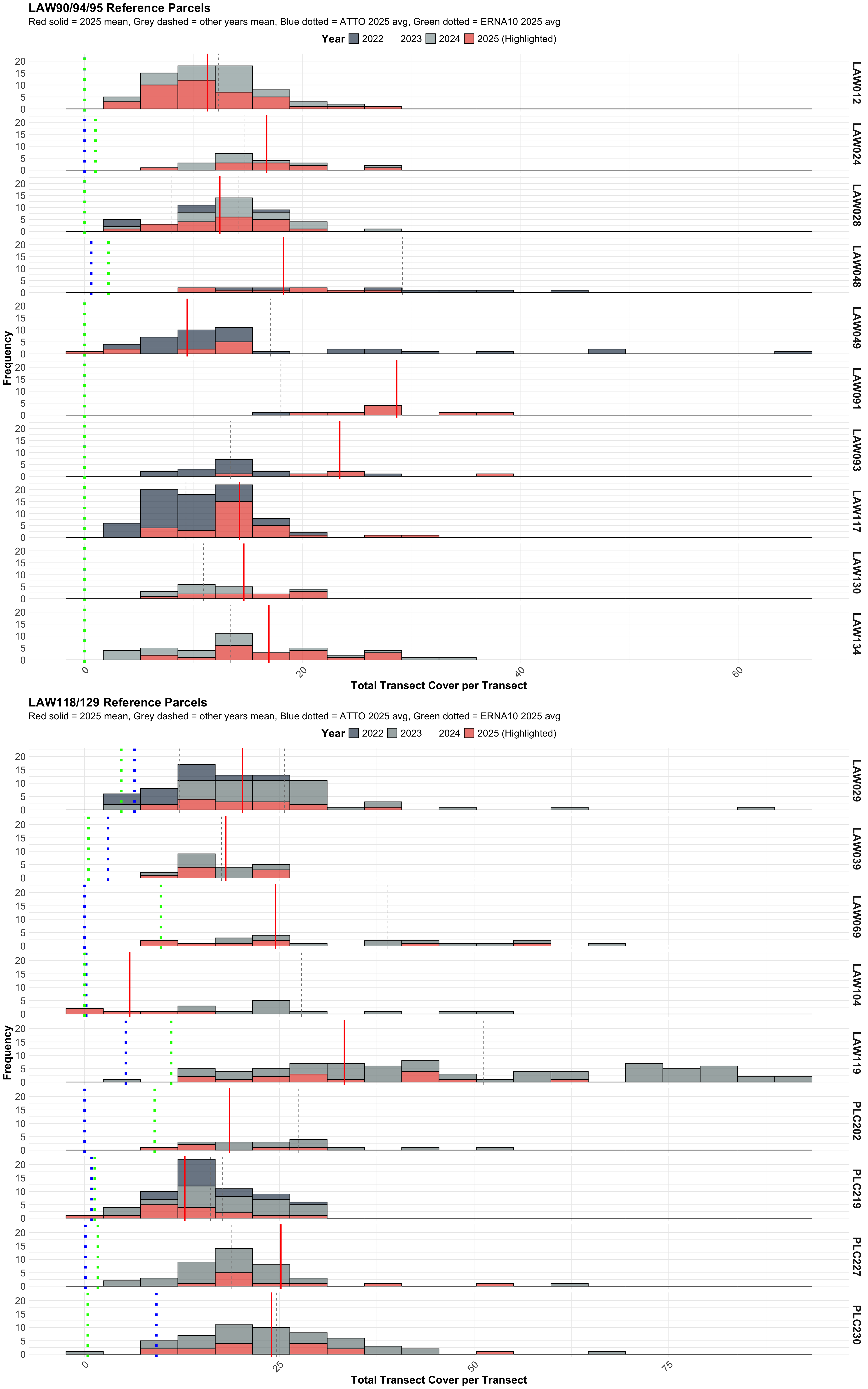
--- title: "Reference Parcels" tbl-cap-location: top affiliation: "Inyo County Water Department" date: "2025-09-03" date-modified: "2025-09-03" citation: type: report container-title: "Data Report" publisher: "Inyo County Water Department" issued: "2025-09-03" url: https://github.com/inyo-gov/revegetation-projects google-scholar: true execute: echo: false warning: false message: false --- ## Data Processing Methodology ### 1. **Data Ingestion** - **LAW90/94/95 parcels**: Processed from individual CSV files- **LAW118/129 parcels**: Processed from Excel files- **Transect normalization**: All transect labels converted to lowercase (e.g., "3A" → "3a")### 2. **Cover Calculation** - **Per-transect basis**: Cover calculated for each transect individually- **Species aggregation**: Multiple hits per species per transect are summed- **Percent cover**: Reference parcel hits directly represent percent cover### 3. **Complete Data Accounting** - **Complete dataset**: All parcel-year-transect-species combinations included- **Zero-filling**: Missing species receive 0% cover (not excluded from averages)- **Group averages**: Averages calculated from ALL parcels in reference groups, including those with zero ATTO/ERNA cover## Reference Parcels Summary ```{r reference-parcels-summary, echo=FALSE, warning=FALSE} #| tbl-cap: "Reference parcels transect counts by group and year." library (dplyr)library (DT)library (readr)library (janitor)library (readxl)library (purrr)library (stringr)library (tidyr)# Load reference data directly <- bind_rows (# LAW90/94/95 reference data (2022-2024) read_csv ("data/raw/reference/law090_094_095_reference_parcel_long_format.csv" , show_col_types = FALSE ) %>% clean_names () %>% select (- date, - green, - live, - x1) %>% rename (transect = tran_name, hits = hit) %>% mutate (transect = as.character (transect)) %>% left_join (read_csv ("data/species.csv" , show_col_types = FALSE ), by = c ("species" = "Code" )) %>% mutate (percent_cover = hits) %>% # Reference parcels: hits = percent cover select (parcel, transect, species, hits, year, CommonName, Lifecycle, Lifeform, percent_cover),# LAW118/129 reference data (2022-2023) read_csv ("data/raw/reference/law118_129_reference_parcel_long_format.csv" , show_col_types = FALSE ) %>% clean_names () %>% select (- date, - green, - live, - x1) %>% rename (transect = tran_name, hits = hit) %>% mutate (transect = as.character (transect)) %>% left_join (read_csv ("data/species.csv" , show_col_types = FALSE ), by = c ("species" = "Code" )) %>% mutate (percent_cover = hits) %>% # Reference parcels: hits = percent cover select (parcel, transect, species, hits, year, CommonName, Lifecycle, Lifeform, percent_cover),# LAW90/94/95 reference data (2025) - from Excel file # Read from ReferenceParcelData sheet (matches GitHub version) read_excel ("data/raw/reveg/LADWP_ReferenceParcel_LAW090_094_095_2025_Data_2025.xlsx" , sheet = "ReferenceParcelData" ) %>% mutate (year = as.integer (Year),transect = as.character (Transect),hits = as.integer (Green + Live),parcel = Parcel,species = Species%>% left_join (read_csv ("data/species.csv" , show_col_types = FALSE ), by = c ("species" = "Code" )) %>% mutate (percent_cover = hits) %>% # Reference parcels: hits = percent cover select (parcel, transect, species, hits, year, CommonName, Lifecycle, Lifeform, percent_cover)# LAW118/129 reference data (2025) - from Excel file # Source the function to read Excel data source ("code/read_reference_parcels_excel.R" )read_reference_parcels_excel ("data/raw/reference/Reference Parcels_LAW_PLC_2025.xlsx" ) %>% left_join (read_csv ("data/species.csv" , show_col_types = FALSE ), by = c ("species" = "Code" )) %>% mutate (percent_cover = hits) %>% # Reference parcels: hits = percent cover select (parcel, transect, species, hits, year, CommonName, Lifecycle, Lifeform, percent_cover)# Create reference parcels summary <- c ('LAW012' , 'LAW024' , 'LAW028' , 'LAW048' , 'LAW049' , 'LAW091' , 'LAW093' , 'LAW117' , 'LAW130' , 'LAW134' )<- c ('LAW029' , 'LAW039' , 'LAW069' , 'LAW104' , 'LAW119' , 'PLC202' , 'PLC219' , 'PLC227' , 'PLC230' )<- ref_data_combined %>% select (parcel, year, transect) %>% distinct () %>% mutate (reference_group = case_when (%in% law90_94_95_parcels ~ 'LAW90/94/95' ,%in% law118_129_parcels ~ 'LAW118/129' ,TRUE ~ 'Other' %>% filter (reference_group != 'Other' ) %>% group_by (reference_group, year) %>% summarise (n_parcels = n_distinct (parcel),n_transects = n (),.groups = 'drop' %>% arrange (reference_group, year) %>% mutate (reference_group = case_when (== 'LAW90/94/95' ~ 'LAW90/94/95' ,== 'LAW118/129' ~ 'LAW118/129' ,TRUE ~ reference_group# Display the table datatable (options = list (autoWidth = TRUE ,scrollX = FALSE ,dom = 'Bfrtip' ,buttons = c ('csv' ),pageLength = 10 rownames = FALSE ,extensions = 'Buttons' ,filter = 'top' ,width = '100%' ,escape = FALSE ,colnames = c ("Reference<br/>Group" ,"Year" ,"Number of<br/>Parcels" ,"Number of<br/>Transects" ``` ```{r setup, message=FALSE, warning=FALSE, echo=FALSE} library (sf)library (here)library (dplyr)library (readr)library (stringr)library (tidyr)library (DT)library (janitor)library (ggplot2)``` ## ```{r species-analysis, warning=FALSE} #| tbl-cap: "Species contribution to perennial cover in reference parcels by year. Shows average cover per transect, average perennial cover per transect, relative contribution percentage, and total transects per parcel-year. Default view filtered to show ATTO and ERNA10 species." #| column: screen # Reference data already loaded in previous chunk # Create comprehensive species analysis with proper zero-inflation handling # For each parcel-year, calculate average cover for each species across ALL transects <- ref_data_combined %>% filter (Lifecycle == 'Perennial' ) %>% # Get all unique combinations of parcel, year, transect, species # But only complete within existing parcel-year combinations group_by (parcel, year) %>% complete (transect, species, fill = list (hits = 0 )) %>% ungroup () %>% # Filter to only include perennial species (since complete() adds all species) filter (species %in% (ref_data_combined %>% filter (Lifecycle == 'Perennial' ) %>% pull (species) %>% unique ())) %>% group_by (parcel, year, species) %>% summarise (avg_cover = mean (hits, na.rm = TRUE ), # Average across all transects (including zeros) n_transects = n_distinct (transect),.groups = 'drop' %>% # Calculate average perennial cover per parcel-year left_join (%>% filter (Lifecycle == 'Perennial' ) %>% group_by (parcel, year, transect) %>% summarise (transect_perennial_cover = sum (hits, na.rm = TRUE ), .groups = 'drop' ) %>% group_by (parcel, year) %>% summarise (avg_perennial_cover = mean (transect_perennial_cover, na.rm = TRUE ), .groups = 'drop' ),by = c ('parcel' , 'year' )%>% # Calculate relative contribution mutate (avg_relative_cover = (avg_cover / avg_perennial_cover) * 100 ) %>% # Get true total transect counts per parcel-year left_join (%>% filter (Lifecycle == 'Perennial' ) %>% group_by (parcel, year) %>% summarise (total_transects = n_distinct (transect), .groups = 'drop' ),by = c ('parcel' , 'year' )%>% # Add reference group column mutate (reference_group = case_when (%in% c ("LAW012" , "LAW024" , "LAW028" , "LAW048" , "LAW049" , "LAW091" , "LAW093" , "LAW117" , "LAW130" , "LAW134" ) ~ "LAW90/94/95" ,%in% c ("LAW029" , "LAW039" , "LAW069" , "LAW104" , "LAW119" , "PLC202" , "PLC219" , "PLC227" , "PLC230" ) ~ "LW118/129" ,TRUE ~ "Other" %>% # Ensure complete zero-filling at parcel level for all species # Create complete grid of all parcel-year-species combinations group_by (reference_group, year, species) %>% complete (parcel = c (# LAW90/94/95 parcels "LAW012" , "LAW024" , "LAW028" , "LAW048" , "LAW049" , "LAW091" , "LAW093" , "LAW117" , "LAW130" , "LAW134" ,# LAW118/129 parcels "LAW029" , "LAW039" , "LAW069" , "LAW104" , "LAW119" , "PLC202" , "PLC219" , "PLC227" , "PLC230" fill = list (avg_cover = 0 , avg_perennial_cover = 0 , avg_relative_cover = 0 , total_transects = 0 ,n_transects = 0 %>% ungroup () %>% # Filter to only include parcels that belong to the current reference group filter (== "LAW90/94/95" & parcel %in% c ("LAW012" , "LAW024" , "LAW028" , "LAW048" , "LAW049" , "LAW091" , "LAW093" , "LAW117" , "LAW130" , "LAW134" )) | == "LW118/129" & parcel %in% c ("LAW029" , "LAW039" , "LAW069" , "LAW104" , "LAW119" , "PLC202" , "PLC219" , "PLC227" , "PLC230" ))%>% # Round numeric columns to 1 decimal place mutate (avg_cover = round (avg_cover, 1 ),avg_perennial_cover = round (avg_perennial_cover, 1 ),avg_relative_cover = round (avg_relative_cover, 1 )%>% # Reorder columns with group first select (reference_group, parcel, year, species, avg_cover, avg_perennial_cover, avg_relative_cover, total_transects) %>% arrange (reference_group, parcel, year, species)# Display the comprehensive species analysis %>% datatable (options = list (autoWidth = TRUE ,scrollX = FALSE ,columnDefs = list (list (width = '15%' , targets = 0 ), # reference_group list (width = '12%' , targets = 1 ), # parcel list (width = '8%' , targets = 2 ), # year list (width = '12%' , targets = 3 ), # species list (width = '12%' , targets = 4 ), # avg_cover list (width = '20%' , targets = 5 ), # avg_perennial_cover (more space) list (width = '20%' , targets = 6 ), # avg_relative_cover (more space) list (width = '15%' , targets = 7 ) # total_transects dom = 'Blfrtip' ,buttons = c ('csv' ),pageLength = 5 ,lengthMenu = list (c (5 , 10 , 20 ), c ('5' , '10' , '20' )),callback = JS (" function(settings) { // Set default filter for Average Cover % column (index 4) to show only non-zero values var api = new $.fn.dataTable.Api(settings); api.column(4).search('>0'); api.draw(); } " )extensions = 'Buttons' ,filter = 'top' ,width = '100%' ,escape = FALSE , # Allow HTML in column names rownames = FALSE , # Remove row numbers to save space colnames = c ("Reference<br/>Parcel Group" ,"Parcel<br/>ID" ,"Year" ,"Species<br/>Code" ,"Average<br/>Cover %" ,"Average<br/>Perennial Cover %" ,"Average<br/>Relative Cover %" ,"Total<br/>Transects" caption = "All species data available. Use filters to view specific species (e.g., ATTO, ERNA10) and years (e.g., 2025)." ,callback = JS (" $('.dataTables_wrapper').css('width', '100%'); $('.dataTables_wrapper').css('overflow-x', 'visible'); " )``` ### ATTO/ERNA10 Summary Statistics ```{r atto-erna-summary, warning=FALSE} #| tbl-cap: "Summary statistics for ATTO and ERNA contribution to reference parcel perennial cover, grouped by species, reference parcel group, and year. Shows mean average cover, standard deviation of average cover, mean relative contribution, and number of parcels per group-year combination." #| column: screen # Create reference parcel group identifier for ATTO/ERNA species only # But include ALL parcels for each reference group-year combination <- species_analysis %>% filter (species %in% c ("ATTO" , "ERNA10" )) %>% # Ensure we have all parcels for each reference group-year combination # by creating a complete grid of reference group × year × species group_by (reference_group, year, species) %>% complete (parcel = c (# LAW90/94/95 parcels "LAW012" , "LAW024" , "LAW028" , "LAW048" , "LAW049" , "LAW091" , "LAW093" , "LAW117" , "LAW130" , "LAW134" ,# LAW118/129 parcels "LAW029" , "LAW039" , "LAW069" , "LAW104" , "LAW119" , "PLC202" , "PLC219" , "PLC227" , "PLC230" fill = list (avg_cover = 0 , avg_perennial_cover = 0 , avg_relative_cover = 0 , total_transects = 0 )) %>% ungroup () %>% # Filter to only include parcels that belong to the current reference group filter (== "LAW90/94/95" & parcel %in% c ("LAW012" , "LAW024" , "LAW028" , "LAW048" , "LAW049" , "LAW091" , "LAW093" , "LAW117" , "LAW130" , "LAW134" )) | == "LW118/129" & parcel %in% c ("LAW029" , "LAW039" , "LAW069" , "LAW104" , "LAW119" , "PLC202" , "PLC219" , "PLC227" , "PLC230" ))# Calculate group-level perennial cover (same for all species in a group-year) <- atto_erna_with_groups %>% group_by (reference_group, year) %>% summarise (group_mean_perennial_cover = mean (avg_perennial_cover, na.rm = TRUE ),n_parcels = n_distinct (parcel),.groups = 'drop' # Calculate summary statistics grouped by species, reference group, AND year <- atto_erna_with_groups %>% group_by (species, reference_group, year) %>% summarise (# Average cover statistics (keep only mean and SD) mean_avg_cover = mean (avg_cover, na.rm = TRUE ),sd_avg_cover = sd (avg_cover, na.rm = TRUE ),# Relative cover statistics (keep only mean) mean_relative_cover = mean (avg_relative_cover, na.rm = TRUE ),.groups = 'drop' %>% # Join with group-level perennial cover left_join (group_perennial_cover, by = c ("reference_group" , "year" )) %>% filter (reference_group != "Other" ) %>% # Remove any "Other" groups # Calculate proportion of species cover relative to total perennial cover mutate (species_proportion_of_total = round ((mean_avg_cover / group_mean_perennial_cover) * 100 , 1 )%>% mutate (across (where (is.numeric), round, 2 )) %>% # Reorder columns for better display (removed species_proportion_of_total as it's redundant with mean_relative_cover) select (species, reference_group, year, mean_avg_cover, sd_avg_cover, group_mean_perennial_cover, mean_relative_cover, n_parcels)# Display summary table %>% datatable (options = list (autoWidth = TRUE ,scrollX = FALSE ,columnDefs = list (list (width = '15%' , targets = 0 ), # species list (width = '18%' , targets = 1 ), # reference_group list (width = '10%' , targets = 2 ), # year list (width = '18%' , targets = 3 ), # mean_avg_cover list (width = '15%' , targets = 4 ), # sd_avg_cover list (width = '18%' , targets = 5 ), # mean_perennial_cover list (width = '18%' , targets = 6 ), # mean_relative_cover list (width = '15%' , targets = 7 ) # n_parcels dom = 'Blfrtip' ,buttons = c ('csv' ),pageLength = 5 ,lengthMenu = list (c (5 , 10 , 20 ), c ('5' , '10' , '20' ))extensions = 'Buttons' ,filter = 'top' ,width = '100%' ,escape = FALSE , # Allow HTML in column names rownames = FALSE , # Remove row numbers to save space colnames = c ("Species<br/>Code" ,"Reference<br/>Parcel Group" ,"Year" ,"Group Average<br/>Species Cover %" ,"Standard<br/>Deviation" ,"Group Average<br/>Total Perennial Cover %" ,"Group Average<br/>Relative Cover %" ,"Number of<br/>Parcels" callback = JS (" $('.dataTables_wrapper').css('width', '100%'); $('.dataTables_wrapper').css('overflow-x', 'visible'); " )``` ```{r reference-parcels-histogram, warning=FALSE} #| fig-cap: "Distribution of total transect cover by reference parcel and year. Two separate plots: one for LAW90/94/95 group and one for LAW118/129 group. Each panel shows one parcel with all years overlaid. 2025 data highlighted in red with red mean line; other years in grey with grey mean lines; blue and green thick lines show ATTO and ERNA10 average cover." #| fig-width: 20 #| fig-height: 32 # Reference data already loaded in previous chunk # Create transect summary for histogram (grouped by parcel, year, and transect) <- ref_data_combined %>% group_by (parcel, year, transect) %>% summarise (Total_Cover = sum (hits, na.rm = TRUE ), .groups = 'drop' ) %>% # Add reference group identifier mutate (reference_group = case_when (%in% c ("LAW012" , "LAW024" , "LAW028" , "LAW048" , "LAW049" , "LAW091" , "LAW093" , "LAW117" , "LAW130" , "LAW134" ) ~ "LAW90/94/95" ,%in% c ("LAW029" , "LAW039" , "LAW069" , "LAW104" , "LAW119" , "PLC202" , "PLC219" , "PLC227" , "PLC230" ) ~ "LW118/129" ,TRUE ~ "Other" # Ensure year is a factor with all levels year = factor (year, levels = c (2022 , 2023 , 2024 , 2025 ))%>% filter (reference_group != "Other" )# Calculate summary statistics by parcel and year for reference lines <- transect_summary %>% group_by (parcel, year, reference_group) %>% summarise (Mean_Cover = mean (Total_Cover, na.rm = TRUE ),SD_Cover = sd (Total_Cover, na.rm = TRUE ),.groups = 'drop' # Calculate overall mean cover per parcel for sorting <- transect_summary %>% group_by (parcel, reference_group) %>% summarise (Overall_Mean_Cover = mean (Total_Cover, na.rm = TRUE ), .groups = 'drop' ) %>% arrange (Overall_Mean_Cover)# Create ordered parcel factor for both groups <- parcel_means %>% filter (reference_group == "LAW90/94/95" ) %>% pull (parcel)<- parcel_means %>% filter (reference_group == "LW118/129" ) %>% pull (parcel)# Apply ordering to transect_summary <- transect_summary %>% mutate (parcel = case_when (== "LAW90/94/95" ~ factor (parcel, levels = law909495_order),== "LW118/129" ~ factor (parcel, levels = law118129_order),TRUE ~ factor (parcel)# Get ATTO and ERNA10 average cover for 2025 only (from species analysis) <- species_analysis %>% filter (species %in% c ("ATTO" , "ERNA10" ), year == 2025 ) %>% select (parcel, year, species, avg_cover) %>% pivot_wider (names_from = species, values_from = avg_cover, names_prefix = "avg_" )# Ensure columns exist even if no data if (! "avg_ATTO" %in% names (atto_erna_cover)) {$ avg_ATTO <- 0 if (! "avg_ERNA10" %in% names (atto_erna_cover)) {$ avg_ERNA10 <- 0 # Replace NA values with 0 <- atto_erna_cover %>% mutate (avg_ATTO = ifelse (is.na (avg_ATTO), 0 , avg_ATTO),avg_ERNA10 = ifelse (is.na (avg_ERNA10), 0 , avg_ERNA10)# Create two separate plots with ATTO/ERNA10 reference lines library (gridExtra)# Plot 1: LAW90/94/95 group <- ggplot (transect_summary %>% filter (reference_group == "LAW90/94/95" ), aes (x = Total_Cover, fill = year)) + geom_histogram (bins = 20 , alpha = 0.7 , color = "black" ) + # Add mean line for 2025 only (in red) geom_vline (data = parcel_year_summary %>% filter (year == 2025 , reference_group == "LAW90/94/95" ), aes (xintercept = Mean_Cover), color = "red" , linetype = "solid" , size = 1 ) + # Add mean line for other years (in grey) geom_vline (data = parcel_year_summary %>% filter (year != 2025 , reference_group == "LAW90/94/95" ), aes (xintercept = Mean_Cover), color = "grey50" , linetype = "dashed" , size = 0.5 ) + # Add ATTO average cover line (in blue) geom_vline (data = atto_erna_cover %>% filter (parcel %in% c ("LAW012" , "LAW024" , "LAW028" , "LAW048" , "LAW049" , "LAW091" , "LAW093" , "LAW117" , "LAW130" , "LAW134" )) %>% filter (avg_ATTO > 0 ),aes (xintercept = avg_ATTO), color = "blue" , linetype = "dotted" , size = 2 ) + # Add ERNA10 average cover line (in green) geom_vline (data = atto_erna_cover %>% filter (parcel %in% c ("LAW012" , "LAW024" , "LAW028" , "LAW048" , "LAW049" , "LAW091" , "LAW093" , "LAW117" , "LAW130" , "LAW134" )) %>% filter (avg_ERNA10 > 0 ),aes (xintercept = avg_ERNA10), color = "green" , linetype = "dotted" , size = 2 ) + labs (title = "LAW90/94/95 Reference Parcels" ,subtitle = "Red solid = 2025 mean, Grey dashed = other years mean, Blue dotted = ATTO 2025 avg, Green dotted = ERNA10 2025 avg" ,x = "Total Transect Cover per Transect" ,y = "Frequency" + facet_grid (parcel ~ ., scales = "free_x" ) + scale_fill_manual (name = "Year" ,values = c ("2022" = "#34495E" , "2023" = "#7F8C8D" , "2024" = "#95A5A6" , "2025" = "#E74C3C" ),labels = c ("2022" , "2023" , "2024" , "2025 (Highlighted)" ),drop = FALSE + theme_minimal () + theme (strip.text = element_text (size = 18 , face = "bold" ),axis.text.x = element_text (angle = 45 , hjust = 1 , size = 16 ),axis.text.y = element_text (size = 16 ),axis.title = element_text (size = 18 , face = "bold" ),plot.title = element_text (size = 20 , face = "bold" ),plot.subtitle = element_text (size = 16 ),legend.text = element_text (size = 16 ),legend.title = element_text (size = 18 , face = "bold" ),legend.position = "top" # Plot 2: LAW118/129 group <- ggplot (transect_summary %>% filter (reference_group == "LW118/129" ), aes (x = Total_Cover, fill = year)) + geom_histogram (bins = 20 , alpha = 0.7 , color = "black" ) + # Add mean line for 2025 only (in red) geom_vline (data = parcel_year_summary %>% filter (year == 2025 , reference_group == "LW118/129" ), aes (xintercept = Mean_Cover), color = "red" , linetype = "solid" , size = 1 ) + # Add mean line for other years (in grey) geom_vline (data = parcel_year_summary %>% filter (year != 2025 , reference_group == "LW118/129" ), aes (xintercept = Mean_Cover), color = "grey50" , linetype = "dashed" , size = 0.5 ) + # Add ATTO average cover line (in blue) geom_vline (data = atto_erna_cover %>% filter (parcel %in% c ("LAW029" , "LAW039" , "LAW069" , "LAW104" , "LAW119" , "PLC202" , "PLC219" , "PLC227" , "PLC230" )) %>% filter (avg_ATTO > 0 ),aes (xintercept = avg_ATTO), color = "blue" , linetype = "dotted" , size = 2 ) + # Add ERNA10 average cover line (in green) geom_vline (data = atto_erna_cover %>% filter (parcel %in% c ("LAW029" , "LAW039" , "LAW069" , "LAW104" , "LAW119" , "PLC202" , "PLC219" , "PLC227" , "PLC230" )) %>% filter (avg_ERNA10 > 0 ),aes (xintercept = avg_ERNA10), color = "green" , linetype = "dotted" , size = 2 ) + labs (title = "LAW118/129 Reference Parcels" ,subtitle = "Red solid = 2025 mean, Grey dashed = other years mean, Blue dotted = ATTO 2025 avg, Green dotted = ERNA10 2025 avg" ,x = "Total Transect Cover per Transect" ,y = "Frequency" + facet_grid (parcel ~ ., scales = "free_x" ) + scale_fill_manual (name = "Year" ,values = c ("2022" = "#34495E" , "2023" = "#7F8C8D" , "2024" = "#95A5A6" , "2025" = "#E74C3C" ),labels = c ("2022" , "2023" , "2024" , "2025 (Highlighted)" ),drop = FALSE + theme_minimal () + theme (strip.text = element_text (size = 18 , face = "bold" ),axis.text.x = element_text (angle = 45 , hjust = 1 , size = 16 ),axis.text.y = element_text (size = 16 ),axis.title = element_text (size = 18 , face = "bold" ),plot.title = element_text (size = 20 , face = "bold" ),plot.subtitle = element_text (size = 16 ),legend.text = element_text (size = 16 ),legend.title = element_text (size = 18 , face = "bold" ),legend.position = "top" # Combine the two plots grid.arrange (plot1, plot2, ncol = 1 )``` ## Reference Data Downloads ```{r reference-data-downloads, warning=FALSE} #| column: screen # Reference data already loaded in previous chunk # Create summary data directly <- ref_data_combined %>% filter (Lifecycle == 'Perennial' ) %>% group_by (parcel, year) %>% summarise (n_transects = n_distinct (transect),total_hits = sum (hits, na.rm = TRUE ),avg_cover = mean (hits, na.rm = TRUE ),.groups = 'drop' %>% mutate (reference_group = case_when (%in% c ('LAW012' , 'LAW024' , 'LAW028' , 'LAW048' , 'LAW049' , 'LAW091' , 'LAW093' , 'LAW117' , 'LAW130' , 'LAW134' ) ~ 'LAW90/94/95' ,%in% c ('LAW029' , 'LAW039' , 'LAW069' , 'LAW104' , 'LAW119' , 'PLC202' , 'PLC219' , 'PLC227' , 'PLC230' ) ~ 'LAW118/129' ,TRUE ~ 'Other' %>% filter (reference_group != 'Other' )# Create ATTO/ERNA analysis <- ref_data_combined %>% filter (species %in% c ("ATTO" , "ERNA10" ), Lifecycle == 'Perennial' ) %>% group_by (parcel, year, species) %>% summarise (avg_cover = mean (hits, na.rm = TRUE ),n_transects = n_distinct (transect),.groups = 'drop' %>% mutate (reference_group = case_when (%in% c ('LAW012' , 'LAW024' , 'LAW028' , 'LAW048' , 'LAW049' , 'LAW091' , 'LAW093' , 'LAW117' , 'LAW130' , 'LAW134' ) ~ 'LAW90/94/95' ,%in% c ('LAW029' , 'LAW039' , 'LAW069' , 'LAW104' , 'LAW119' , 'PLC202' , 'PLC219' , 'PLC227' , 'PLC230' ) ~ 'LAW118/129' ,TRUE ~ 'Other' %>% filter (reference_group != 'Other' )# Save datasets for download write_csv (reference_parcel_summary, "data/processed/reference_parcel_summary.csv" )write_csv (reference_atto_erna_analysis, "data/processed/reference_atto_erna_analysis.csv" )write_csv (ref_data_combined, "data/processed/ref_data_combined.csv" )# Function to get file modification date <- function (file_path) {if (file.exists (file_path)) {<- file.info (file_path)return (format (file_info$ mtime, "%Y-%m-%d %H:%M" ))else {return ("File not found" )# File paths for the reference data downloads <- c ("data/processed/reference_parcel_summary.csv" ,"data/processed/reference_atto_erna_analysis.csv" ,"data/processed/ref_data_combined.csv" # Create summary table of available datasets with download links and modification dates <- data.frame (Dataset = c ("Reference Parcel Summary" ,"ATTO/ERNA Analysis" ,"Combined Reference Data" Description = c ("Summary statistics by parcel and year" ,"ATTO and ERNA species analysis by parcel-year" ,"Complete reference dataset with all variables" Rows = c (nrow (reference_parcel_summary),nrow (reference_atto_erna_analysis),nrow (ref_data_combined)Columns = c (ncol (reference_parcel_summary),ncol (reference_atto_erna_analysis),ncol (ref_data_combined)"Last Modified" = unname (sapply (ref_file_paths, get_file_date)),Download = c ('<a href="https://github.com/inyo-gov/revegetation-projects/raw/main/data/processed/reference_parcel_summary.csv" download>⬇ CSV</a>' ,'<a href="https://github.com/inyo-gov/revegetation-projects/raw/main/data/processed/reference_atto_erna_analysis.csv" download>⬇ CSV</a>' ,'<a href="https://github.com/inyo-gov/revegetation-projects/raw/main/data/processed/ref_data_combined.csv" download>⬇ CSV</a>' # Display download information datatable (options = list (autoWidth = TRUE ,scrollX = FALSE ,columnDefs = list (list (width = '20%' , targets = 0 ), # Dataset Name list (width = '33%' , targets = 1 ), # Description list (width = '10%' , targets = 2 ), # Number of Rows list (width = '10%' , targets = 3 ), # Number of Columns list (width = '12%' , targets = 4 ), # Last Modified list (width = '15%' , targets = 5 ) # Download dom = 'Bfrtip' ,buttons = c ('csv' ),pageLength = 10 extensions = 'Buttons' ,filter = 'top' ,width = '100%' ,escape = FALSE , # Allow HTML in column names and Download column rownames = FALSE , # Disable automatic row numbering colnames = c ("Dataset<br/>Name" ,"Description<br/>& Details" , "Number<br/>of Rows" ,"Number<br/>of Columns" ,"Last<br/>Modified" ,"Download<br/>Link" callback = JS (" $('.dataTables_wrapper').css('width', '100%'); $('.dataTables_wrapper').css('overflow-x', 'visible'); $('.dataTables_wrapper td:nth-child(1)').css('white-space', 'normal'); $('.dataTables_wrapper td:nth-child(1)').css('word-wrap', 'break-word'); $('.dataTables_wrapper td:nth-child(1)').css('max-width', '20%'); $('.dataTables_wrapper td:nth-child(2)').css('white-space', 'normal'); $('.dataTables_wrapper td:nth-child(2)').css('word-wrap', 'break-word'); $('.dataTables_wrapper td:nth-child(2)').css('max-width', '33%'); " )``` ## Transects per Reference Parcel by Year ```{r transects-by-parcel-year-fresh, echo=FALSE, cache=FALSE} # Calculate transects per reference parcel by year - FRESH VERSION # Use raw reference data to get ALL transects, not just ATTO/ERNA analysis # Force refresh: 2025-01-10 16:35 library (dplyr)library (tidyr)# Reference data already loaded in previous chunk # Calculate transects per parcel-year <- ref_data_combined %>% filter (Lifecycle == 'Perennial' ) %>% select (parcel, year, transect) %>% distinct () %>% mutate (reference_group = case_when (%in% c ('LAW012' , 'LAW024' , 'LAW028' , 'LAW048' , 'LAW049' , 'LAW091' , 'LAW093' , 'LAW117' , 'LAW130' , 'LAW134' ) ~ 'LAW90/94/95' ,%in% c ('LAW029' , 'LAW039' , 'LAW069' , 'LAW104' , 'LAW119' , 'PLC202' , 'PLC219' , 'PLC227' , 'PLC230' ) ~ 'LAW118/129' ,TRUE ~ 'Other' %>% filter (reference_group != 'Other' ) %>% group_by (reference_group, parcel, year) %>% summarise (n_transects = n (), .groups = 'drop' ) %>% pivot_wider (names_from = year,values_from = n_transects,names_prefix = "Year_" ,values_fill = 0 %>% # Reorder columns to be chronological (only select columns that exist) select (reference_group, parcel, any_of (c ("Year_2022" , "Year_2023" , "Year_2024" , "Year_2025" ))) %>% arrange (reference_group, parcel)# Create the table in the same chunk datatable (options = list (autoWidth = TRUE ,scrollX = TRUE ,dom = 'Bfrtip' ,buttons = c ('csv' ),pageLength = 15 rownames = FALSE ,extensions = 'Buttons' ,filter = 'top' ,width = '100%' ,escape = FALSE ,colnames = c ("Reference<br/>Group" ,"Parcel" ,"2022" ,"2023" , "2024" ,"2025" ```